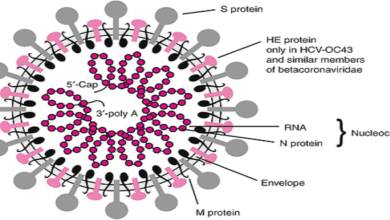
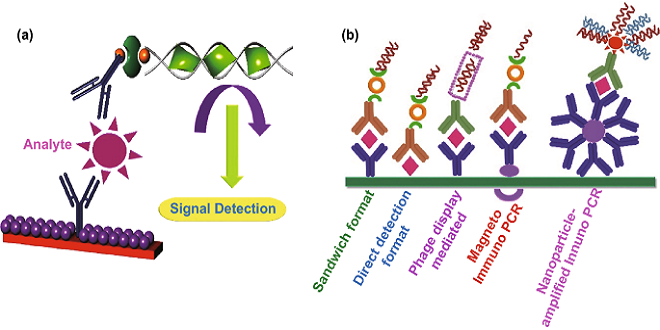
Virus Detection Following Biochemical Amplification
Polymerase chain reaction
. This method provides a highly sensitive test for viral genomes. First, the nucleic acid is extracted from the patient material to be analyzed. Any RNA virus genome present in the material is transcribed into DNA by reverse transcriptase (see p. 385f.).
This DNA, as well as the DNA of the DNA viruses, is then replicated in vitro with a DNA polymerase as follows: after the DNA double-strand has been separated by applying heat, two synthetic oligonucleotides are added that are complementary to the two ends of the viral genome segment being looked for and can hybridize to it accordingly. The adjacent DNA (toward each 50 ends) is then
Two oligonucleotide primers are hybridized to the DNA double strands, which have been separated by heating. A heat-stable polymerase (e.g., Taq polymerase from Thermus aquaticus) is then added and extends these primers along the length of, and complementary to, the matrix strand. The resulting double strands are then once again separated by heat and the reaction is repeated.
b The DNA strands produced in the first cycle (1st generation) have a defined 50 end (corresponding to the primer) and an undefined 30 end. All of the subsequent daughter strands (2nd to nth generation) have a uniform, defined length.
The new and old strands are once again separated by heat and the reaction is started over again. Running several such cycles amplifies the original viral DNA by a factor of many thousands. Beginning with the second generation, the newly synthesized DNA strands show a uniform, defined length and are therefore detectable by means of gel electrophoresis.
The specificity of the reaction is verified by checking the sequences of these DNA strands by means of hybridization or sequencing. The amplification and detection systems in use today for many viruses are increasingly commercially available, and in some cases are also designed to provide quantitative data on the “viral load.”
Serodiagnosis
If a viral infection induces humoral immunity (see p. 48f. and 401), the resulting antibodies can be used in a serodiagnosis. When interpreting the serological data, one is confronted by the problem of deciding whether the observed reactions indicate a fresh, current infection or earlier contact with the virus in question. Two criteria can help with this decision:
Detection of IgM (without IgG) proves the presence of fresh primary infection. IgM is now usually detected by specific serum against human IgM in the so-called capture test, an EIA (p. 128). To test for IgM alone, a blood specimen must be obtained very early in the infection cycle. Concurrent detection of IgG and IgM in blood sampled somewhat later in the course of the disease would also indicate a fresh infection.
Teluguwap offers a diverse range of Telugu music, movies, and more, making it a go-to platform for Telugu entertainment enthusiasts. With its user-friendly interface and vast collection, it provides a convenient and immersive experience. Whether you are a fan of classic melodies or the latest blockbuster hits, Teluguwap has got you covered.
Last word
It could, however, also indicate a reactivated latent infection or an anamnestic reaction (i.e., a nonspecific increase in antibodies in reaction to a nonrelated infection), since IgM can also be produced in both of these cases.
A fourfold increase in the IgG titer within 10–14 days early on in the course of the infection or a drop of the same dimensions later in the course would also be confirmed.